Green infrastructure (GI), utilizing plant or soil systems, permeable pavement, and other mitigators to filter and absorb stormwater where it falls, is a topic of interest to a variety of stakeholders. City and local government officials, utility and engineering companies, and environmental organizations as well as private citizens recognize the need to reduce potentially dangerous and contaminated water flows to sewer systems or surface waters.
Efficient stormwater management in highly urbanized communities is especially critical, considering how floods and droughts could be caused or exacerbated by climate change. However, creating an effective model to examine runoff possibilities requires careful consideration of a wide array of factors including climate data, soil types, water flow patterns, and current land usage. Furthermore, it is imperative that other elements — geographic, demographic, economic, and ecological — be factored into the model.
 That was just the challenge Younsung Kim in Mason’s Department of Environmental Science and Policy wanted to take on. Kim, who has an extensive background in complex environmental and sustainability issues, saw the potential of incorporating freely available data from US Geological Society and Census Bureau along with the local county government land use zoning data to create a computational spatial model to help identify what works, and what doesn’t work, in GI.
That was just the challenge Younsung Kim in Mason’s Department of Environmental Science and Policy wanted to take on. Kim, who has an extensive background in complex environmental and sustainability issues, saw the potential of incorporating freely available data from US Geological Society and Census Bureau along with the local county government land use zoning data to create a computational spatial model to help identify what works, and what doesn’t work, in GI.
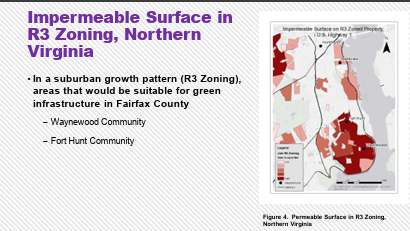
Kim also recognized the importance of applying the prototype in a real-world setting. To do so, she and her research team identified several eastern Fairfax County locations along the Route 1 corridor in Virginia, including the communities of Huntington, Waynewood, and Fort Hunt to illustrate how the model might work.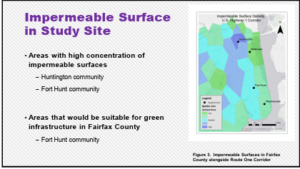 Finally, Kim wanted to incorporate today’s trends in GI architecture in the research; identifying UVA’s School of Architecture as a leading source of urban planning and spatial analysis expertise.
Finally, Kim wanted to incorporate today’s trends in GI architecture in the research; identifying UVA’s School of Architecture as a leading source of urban planning and spatial analysis expertise.
This robust goal was brought to fruition after Kim’s proposal “Assessing Green Infrastructure Potential Using Multi-level Ecological and Economic Factors: The Northern Virginia Case” won a 4-VA@Mason Collaborative Research Grant. Kim’s co-PI at UVA, Vanessa Guerra, received a 4-VA Complementary Grant for her work on the project.
Kim also tapped a number of other sources, including Alex Iszard and Greg Farley in Mason’s Facilities Administration, who offered their GI perspective from a facility management standpoint.
Kim’s PhD student Colin Chadduck assisted in the development of the computational model, studying the ecological and economic factors important for GI site identification. Mason undergraduate students Caroline Miller, Dahvi E Hochman, and Nicolas Bataille provided data collection and literature review support. Undergraduate Sunho Oh played the same role at UVA. Additionally, Elizabeth Grant, formerly a professor at Virginia Tech, advised on the subject of experimental designs of green roof systems.
Together, the team developed a proven successful computational model incorporating ecological and economic dimensions for GI placement which has received national and international attention.
The research outcomes were presented at the American Society for Public Administration Annual Conference, the Midwest Political Science Association Conference, and at a Mason Earth Day event. A paper entitled “Mapping Green Infrastructure from Stormwater” was published in Environmental Pollution and Climate Change.
Political Science Association Conference, and at a Mason Earth Day event. A paper entitled “Mapping Green Infrastructure from Stormwater” was published in Environmental Pollution and Climate Change.
Based on the spatial analysis method developed on the project, Kim won a grant from the Korea International Cooperation Agency to develop resilience enhancement action programs for climate refugee communities in the Philippines.
The project now extends to increase public awareness. Kim and her students are developing a GI webpage that will include resources and a video clip focusing on green infrastructure.
“This 4-VA@Mason grant has created results-oriented direction for GI within Fairfax County, the Commonwealth of Virginia, and beyond; and cemented a long-term collaborative relationship between our environmental policy research group here at Mason and the environmental planning lab at UVA,” noted Kim. “We believe our results have far-reaching possibilities and impact on urban sustainability.”













 recognizes there is much to be understood about creating constructive introductions in the school setting. However, she is also keenly aware of a key flaw in the data used in the benchmark research – it is predominantly limited to students who are economically advantaged.
recognizes there is much to be understood about creating constructive introductions in the school setting. However, she is also keenly aware of a key flaw in the data used in the benchmark research – it is predominantly limited to students who are economically advantaged. Several months later, with that grant in hand, Garner identified an undergrad student, Tamera Toney, who was interested in the project and would be able to handle some of the data entry and management responsibilities. Toney worked on the project during her senior year at Mason. Garner saw that the 4-VA funding could provide a personal and professional pathway for Toney to enhance her studies. Toney recently graduated and will return in the fall to begin her master’s work in Psychology.
Several months later, with that grant in hand, Garner identified an undergrad student, Tamera Toney, who was interested in the project and would be able to handle some of the data entry and management responsibilities. Toney worked on the project during her senior year at Mason. Garner saw that the 4-VA funding could provide a personal and professional pathway for Toney to enhance her studies. Toney recently graduated and will return in the fall to begin her master’s work in Psychology.

 rehearsed and studied the music within its historical context. And similar to good musical composition, RtA worked to a crescendo. For the RtA team, that was a spring evening in Charlottesville when the team of researchers, performers (musicians and singers), videographers and archivists, librarians, faculty and more joined together in UVA’s Colonnade Club Garden Room to fully embrace 17 pieces of WWI music. From “K-K-K Katy” to “Over There” to “Oh How I Hate to Get Up in the Morning” the Colonnade Room sprang to life — circa 1918.
rehearsed and studied the music within its historical context. And similar to good musical composition, RtA worked to a crescendo. For the RtA team, that was a spring evening in Charlottesville when the team of researchers, performers (musicians and singers), videographers and archivists, librarians, faculty and more joined together in UVA’s Colonnade Club Garden Room to fully embrace 17 pieces of WWI music. From “K-K-K Katy” to “Over There” to “Oh How I Hate to Get Up in the Morning” the Colonnade Room sprang to life — circa 1918.


 Team member Psyche Z. Ready, assisted by Joyce P. Johnston, took the lead in adapting Mason Journals’ iteration of the Open Journal System (OJS) to meet the needs of English 302 OER collection authors, reviewers, and users. Each item in the new, public-facing collection includes an abstract, instructor’s notes, and creative-commons licensed curricular materials – assignments, activities, and/or background readings – created, adapted, or curated for use in English 302. The OJS platform eases the review process, and also allows user-friendly features such as keyword searching.
Team member Psyche Z. Ready, assisted by Joyce P. Johnston, took the lead in adapting Mason Journals’ iteration of the Open Journal System (OJS) to meet the needs of English 302 OER collection authors, reviewers, and users. Each item in the new, public-facing collection includes an abstract, instructor’s notes, and creative-commons licensed curricular materials – assignments, activities, and/or background readings – created, adapted, or curated for use in English 302. The OJS platform eases the review process, and also allows user-friendly features such as keyword searching.